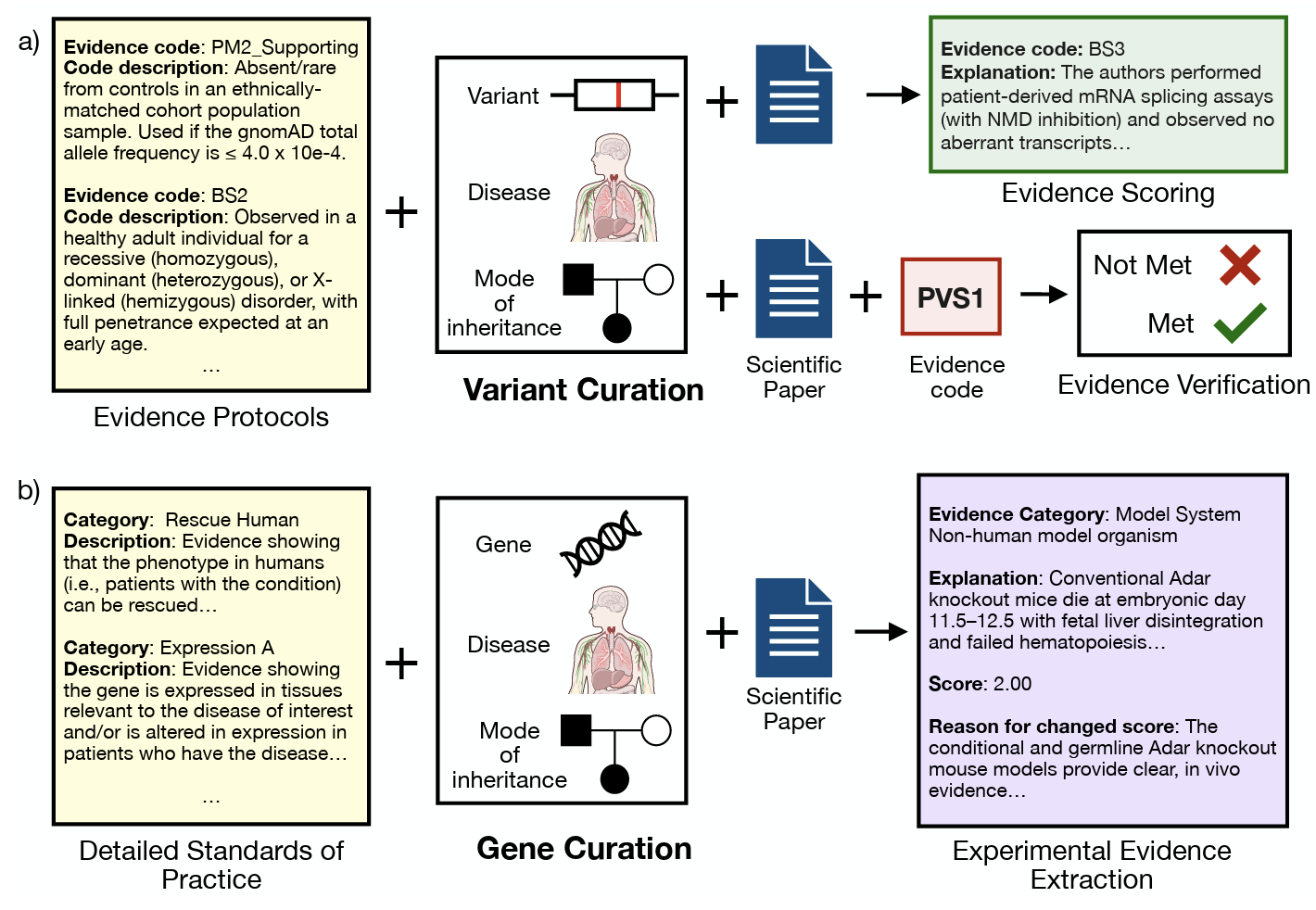
CGBench: Benchmarking Language Model Scientific Reasoning for Clinical Genetics Research
Neural Information Processing Systems (NeurIPS) · 2025
All work
Neural Information Processing Systems (NeurIPS) · 2025

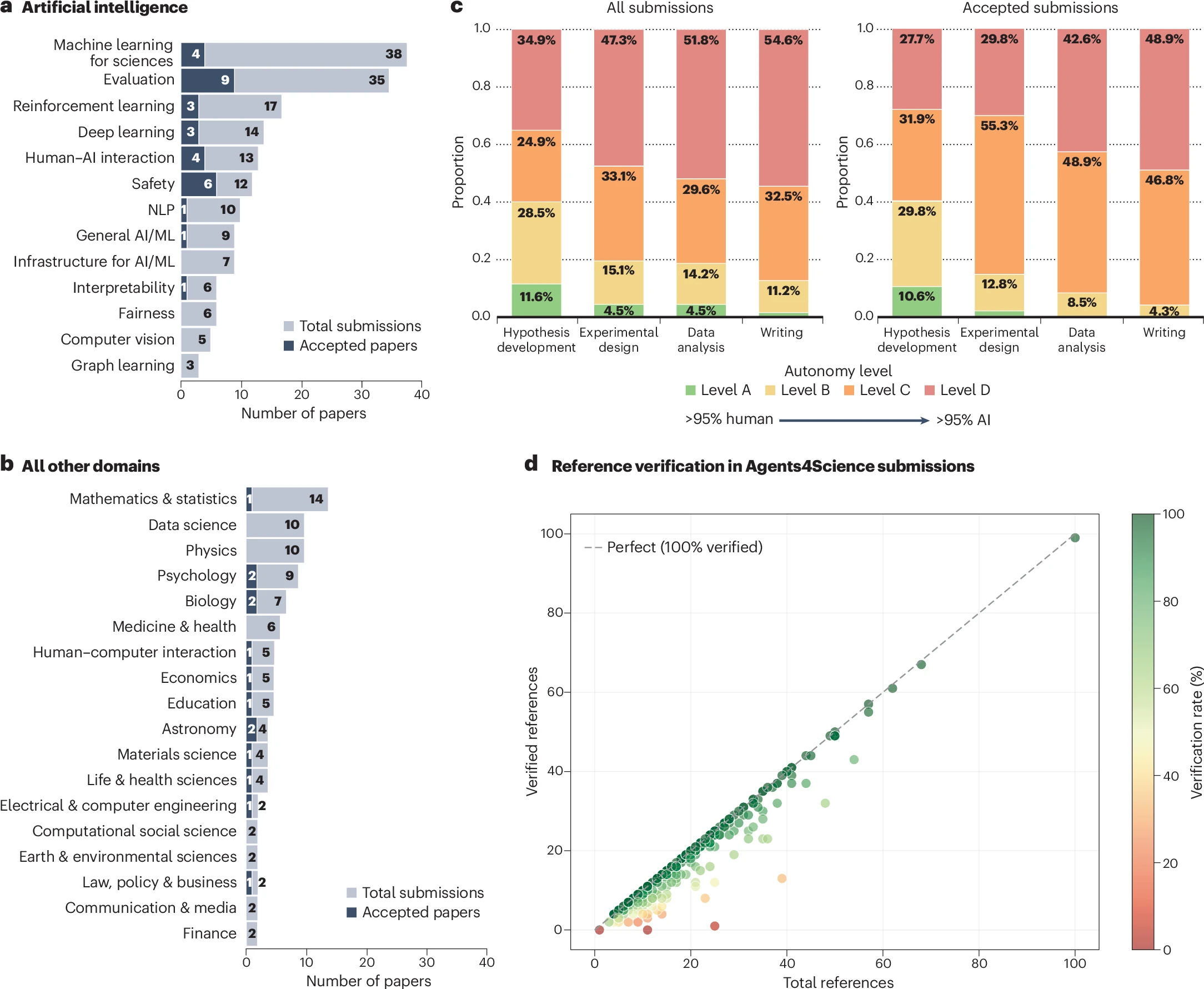
bioRxiv · 2025
ProCyon is an 11B-parameter model that predicts and generates protein phenotypes across molecular and therapeutic scales, enabling transfer to poorly characterized proteins.
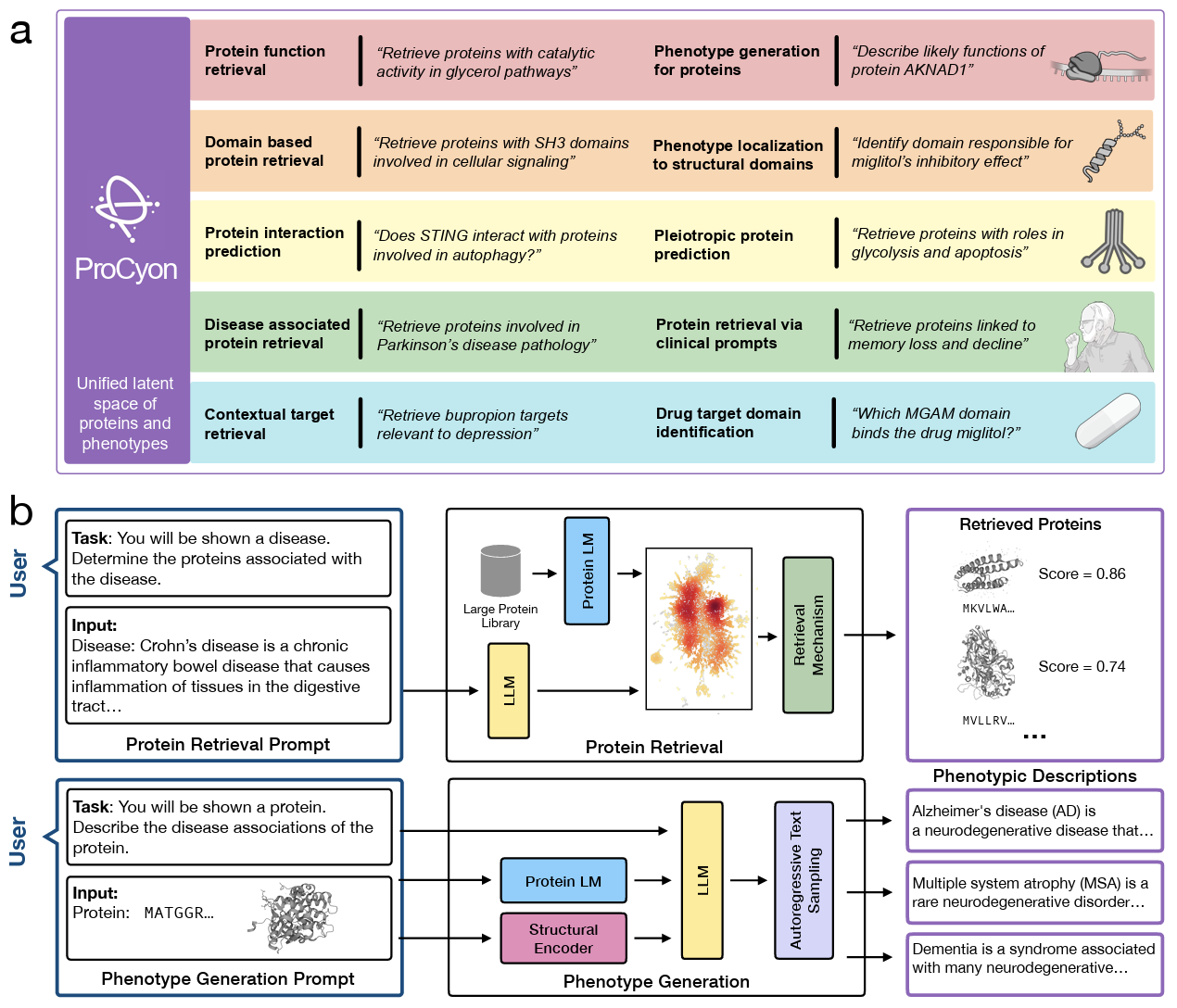
Annual Review of Biomedical Data Science · 2024
We survey graph machine learning for biomedical applications, outlining how relational models and emerging foundation models can deliver clinically meaningful predictions.
International Conference on Machine Learning (ICML) · 2023
RAINCOAT introduces a domain adaptation framework tailored to time series, achieving state-of-the-art performance even when shifts appear in both features and labels.
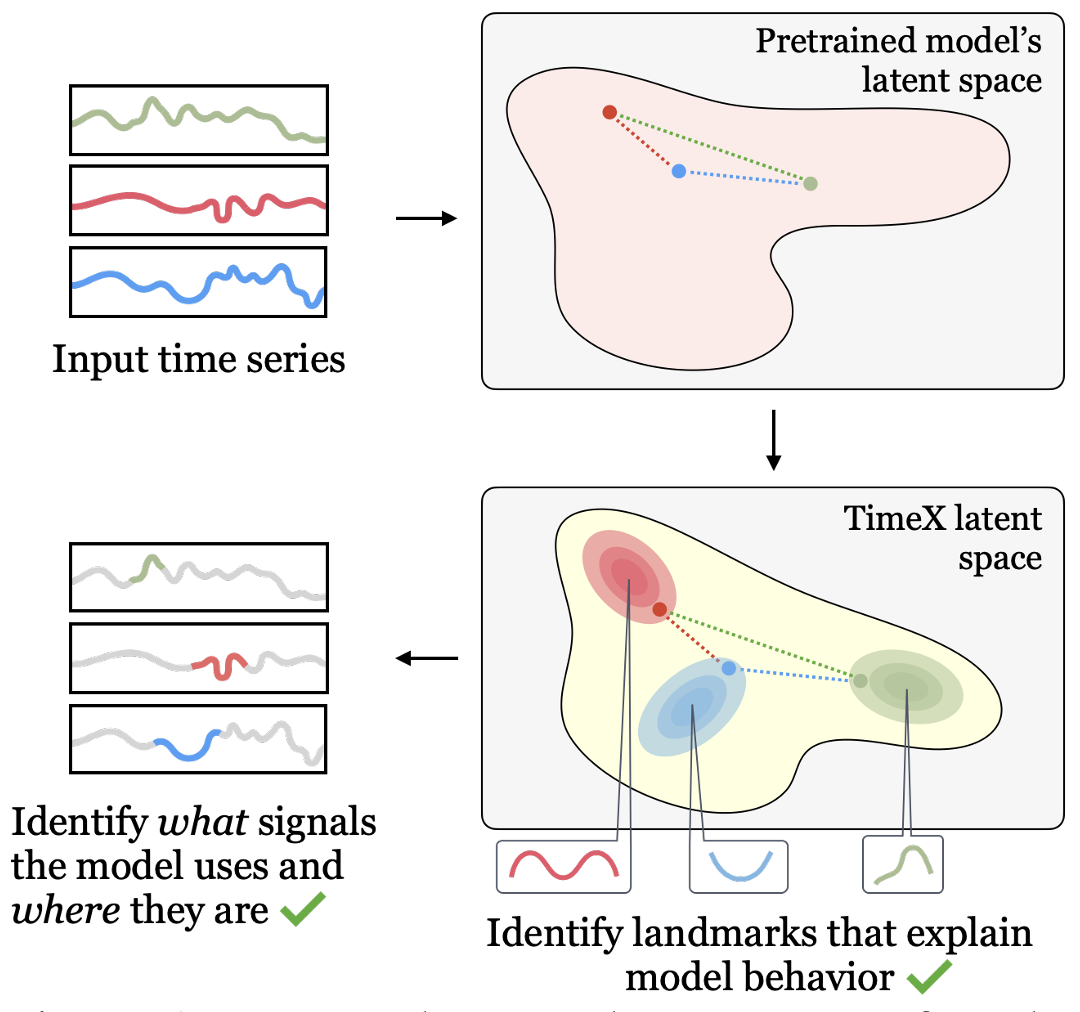
Neural Information Processing Systems (NeurIPS) · 2023
TimeX learns interpretable surrogate masks that mirror predictor behavior through a novel consistency loss, delivering faithful explanations for time-series models.
Scientific Data · 2023
GraphXAI benchmarks GNN explainers with new datasets, metrics, and evaluation flows that now inform tooling such as PyTorch Geometric.
npj Computational Materials · 2023
We design a GNN architecture for polymer property prediction, achieving state-of-the-art performance and enabling high-throughput materials search.
2023 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), DLBIBM Workshop · 2023
We fine-tune protein language models on microbial genomics data and pair them with structure-aware explanations to reveal functional differences between surface and subsurface proteins.
2021 IEEE International Conference on Bioinformatics and Biomedicine (BIBM) · 2021
LASSO-driven feature selection reliably identifies informative genomic features for sepsis prediction and environmental profiling in microbial datasets.